Abstract
Purpose
CTNNA1 is a potential diffuse gastric cancer risk gene, however CTNNA1 testing on multigene panel testing (MGPT) remains unstudied.
Methods
De-identified data from 151,425 individuals who underwent CTNNA1 testing at a commercial laboratory between October 2015 and July 2019 were reviewed. Tissue α-E-catenin immunohistochemistry was performed on CTNNA1 c.1351C>T (p.Arg451*) carriers.
Results
Fifty-two individuals (0.03% tested) had CTNNA1 loss-of-function (LOF) variants and 1057 individuals (0.7% tested) had a total of 302 distinct missense variants of uncertain significance. Detailed history was available on 33 CTNNA1 LOF carriers, with 21 unique CTNNA1 LOF variants. Four (12%) individuals had diffuse gastric cancer and 22 (67%) had breast cancer. Six (21%) and 24 (83%) of the 29 families reported a history of gastric or breast cancer, respectively. The CTNNA1 c.1351C>T nonsense variant was identified in three separate families with early-onset diffuse gastric cancer or breast cancer. Immunohistochemistry showed decreased α-E-catenin expression in gastric cancers.
Conclusion
CTNNA1 LOF variants are detected on MGPT with a majority of these individuals having gastric or breast cancer. The overall risk of gastric cancer for CTNNA1 LOF carriers may be lower than expected. Given the uncertain phenotype and penetrance, management of individuals with CTNNA1 LOF variants remains challenging.
Similar content being viewed by others
INTRODUCTION
With improving technology and accessibility, the use of clinical germline genetic testing for cancer risk assessment has increased significantly.1,2 The most common modality of germline genetic testing for cancer risk assessment is multigene panel testing (MGPT), and the number of genes available for testing via MGPT continues to increase.1,2 Pan-cancer and cancer site–specific MGPT is available for many cancer types including gastric cancer, which remains one of the most common cancers worldwide.3 Based on the Lauren classification gastric cancer is classically divided between intestinal and diffuse types, with diffuse gastric cancer being the less common subtype seen in approximately 30% of cases.4 While up to 20% of individuals diagnosed with gastric cancer have a family history of gastric cancer, only 1–3% of these cancers are thought to be due to a hereditary cancer risk syndrome caused by an identifiable genetic variant.5 One of the best characterized syndromes is hereditary diffuse gastric cancer syndrome (HDGC), which is associated with an increased risk of diffuse gastric cancer as well as lobular breast cancer.6 HDGC often results from germline pathogenic variants in the CDH1 gene, which codes for the cell–cell adhesion protein E-cadherin. Germline testing for CDH1 is recommended currently for individuals meeting HDGC testing criteria.6
The majority of families meeting HDGC criteria do not have a detectable pathogenic CDH1 variant. In 2013, exome sequencing identified a truncating variant in the CTNNA1 gene in a HDGC family with no detectable germline CDH1 variant.7 CTNNA1 codes for α-E-catenin, which is a protein that interacts with E-cadherin. Following this initial report, there have been subsequent publications documenting individuals with diffuse gastric cancer and truncating CTNNA1 variants, highlighting the possibility that CTNNA1 may be a gastric cancer risk gene similar to CDH1.8,9,10,11 However, unlike CDH1, the gastric cancer penetrance and whether CTNNA1 is associated with increased breast cancer risk remain uncertain.
In addition to the unknown cancer penetrance, the appropriate management of individuals with loss-of-function (LOF) CTNNA1 variants remains unclear. Carriers of pathogenic CDH1 variants are typically recommended to undergo risk-reducing total gastrectomy, as gastric cancer screening in CDH1 carriers is not effective.12 While a recent report highlighted the first risk-reducing total gastrectomy performed in a CTNNA1 carrier, there remain no clear guidelines about gastric cancer risk management in this group.8 Much of the uncertainty related to the management of CTNNA1 is due to the limited knowledge about the prevalence and penetrance of CTNNA1 LOF variants, as until recently, commercial genetic testing laboratories did not offer clinical CTNNA1 testing.
Herein, we provide the first large-scale report of CTNNA1 sequencing findings on MGPT through a commercial lab. The data reveal multiple new CTNNA1 LOF variants, including a new truncating variant found in multiple families that may represent a risk factor for early-onset diffuse gastric cancer. These data also provide an important overview of the demographics and cancer history of CTNNA1 LOF variant carriers, which may aid in clinical decision making.
MATERIALS AND METHODS
Patient cohort
Personal and family history information from submitted requisition forms and medical records (when available) were reviewed for 151,425 individuals who underwent germline genetic testing between October 2015 and July 2019. All patient data was de-identified before analysis under Western Institutional Review Board (IRB) protocol number 1167406, which allowed for study and publication of de-identified patient data. Ethnicity was provided by all patients at the time of test ordering and was grouped for analysis based on categories used in population databases.
Genetic testing
Genes analyzed for each patient were chosen at the discretion of the ordering clinician. All MGPT included CTNNA1 and CDH1. Sample types for this cohort included blood and saliva. Once extracted, DNA was processed and subjected to paired-end sequencing on an Illumina next-generation sequencing platform (depth of coverage 50× minimum, 350× average), as described previously.13 Variants were subjected to clinical interpretation using refined American College of Medical Genetics and Genomics criteria (Sherloc).14 All CTNNA1 variants included on clinical genetic testing reports provided to ordering providers were classified as variants of uncertain significance (VUS). CTNNA1 VUSs included CTNNA1 LOF variants, which were defined as variants predicted to lead to protein truncation (nonsense and frameshift variants), large deletions of one or more exons, or consensus splice site variants predicted to disrupt splicing in CTNNA1, as well as CTNNA1 missense VUSs, which were defined as missense variants detected in CTNNA1 that were not classified as benign or likely benign. De-identified variants and their interpretations from this study have been submitted to ClinVar (National Center for Biotechnology Information, Bethesda, MD; http://www.ncbi.nlm.nih.gov/clinvar).
CTNNA1 c.1351C>T families
Probands with the CTNNA1 c.1351C>T variant (p.Arg451*, reference transcript NM_001903.3) were separately consented for enrollment into IRB-approved research protocols at the University of Pennsylvania Perelman School of Medicine (Philadelphia, PA) or City of Hope (Duarte, CA). Once consent was obtained, detailed personal and family history was obtained to allow for pedigree generation. Tissue sections from individuals with gastric adenocarcinoma were acquired, and immunohistochemistry (IHC) of these formalin-fixed paraffin-embedded tissues was performed using an antibody against human α-E-catenin (Thermo Fisher 13–9700, clone alpha-CAT-7A4) using a heat-induced epitope retrieval method. Heat-induced epitope retrieval was performed for 20 minutes with ER2 solution (Leica Microsystems AR9961). Incubation with the α-E-catenin antibody at 1:100 was performed for 15 minutes followed by an 8-minute postprimary step and an 8-minute incubation with polymer HRP. Blocking with endogenous peroxidase for 5 minutes was performed followed by 10 minutes with DAB. Slides were washed three times between each step with bond wash buffer or water. IHC staining was captured on a Leica Bond-IIITM instrument using the Bond Polymer Refine Detection System (Leica Microsystems DS9800).
Statistical analysis
Fisher’s exact test was utilized to calculate p values for the difference between the frequency of breast and gastric cancers in individuals with CTNNA1 LOF variants compared with all individuals tested in the cohort. Significance is defined as p < 0.05.
RESULTS
Over the study period, CTNNA1 sequencing was performed in 151,425 individuals (Fig. 1). Of these 151,425 individuals, 1240 (0.8%) had a personal history of gastric cancer, and 52,093 (34%) had a personal history of breast cancer. 51,185 (34%) had a history of a nongastric, nonbreast cancer, and 46,907 (31%) had no personal history of cancer.
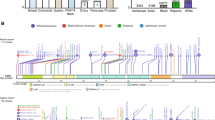
Of the individuals who underwent CTNNA1 sequencing, 52 individuals (0.03% of those tested) had CTNNA1 LOF variants detected. Detailed history was available on 33 individuals with CTNNA1 LOF variants (Table 1), including 2 males (6%) and 31 females (94%). The 33 individuals with detailed history available were from 29 different families, with 4 of these individuals being identified via cascade testing in 2 of the families. Among the 29 different families there were a total of 21 unique CTNNA1 LOF variants, with only CTNNA1 c.2023C>T (p.Gln675*) being previously reported in the literature.8 Of those with LOF variants, all affected probands had negative CDH1 testing, and no individuals had another pathogenic/likely pathogenic variant identified in any high or moderate risk cancer susceptibility gene. Four (12%) individuals had a personal history of diffuse gastric cancer, which was significantly greater than the 0.8% (1240/151,425) of the tested cohort with gastric cancer (p = 0.0002). Twenty-two (67%) individuals had a personal history of invasive breast cancer or ductal carcinoma in situ, excluding 1 individual with known lobular carcinoma in situ (LCIS), which was significantly greater than the 34% (52,093/151,425) of the tested cohort with breast cancer (p = 0.0002). Of the 7 (21%) individuals who were unaffected with cancer, the majority (5 individuals) had a family history of gastric and/or breast cancer in a first-degree relative.
Taking together the probands and first-degree relatives in the 29 families, 6 (21%) families had a history of gastric cancer, 3 (10%) families had a history of gastric and breast cancer, 21 (72%) families had a history of breast cancer without gastric cancer, and 2 (7%) families had no reported history of gastric or breast cancer. In addition to these LOF variants, 1057 individuals (0.7% of those tested) had a CTNNA1 missense VUS identified, representing 302 distinct variants (Supplemental Table 1).
Of particular interest from the LOF variants was the novel CTNNA1 c.1351C>T nonsense variant, which is predicted to lead to a protein truncation product (p.Arg451*). This CTNNA1 variant was identified in three separate families. The proband of family 13 is a Caucasian female who was diagnosed with metastatic, poorly differentiated, diffuse signet-ring cell gastric adenocarcinoma at age 25 (Fig. 2a). Immunohistochemistry (IHC) revealed loss of α-E-catenin expression in the gastric adenocarcinoma with retained α-E-catenin expression in the normal gastric epithelium (Fig. 2b, c). The patient’s mother, who had thyroid cancer at age 47 and LCIS at age 53, as well as her unaffected brother, both tested positive for the CTNNA1 variant and underwent upper endoscopy with Cambridge protocol biopsies and no gross lesions were identified and no foci of signet-ring cells were identified on biopsies. The remainder of the family history is significant for her maternal grandfather with prostate cancer at age 65 and colorectal cancer at age 70, and a maternal first cousin once removed with breast cancer diagnosed at age 50.
a Pedigree of family 13. Red = gastric cancer, yellow = breast cancer and lobular carcinoma in situ (LCIS), gray = other cancers. (b, c) Hematoxylin & eosin (H&E) staining and α-E-catenin immunohistochemistry (IHC) of endoscopic biopsies of the family 13 proband’s diffuse signet-ring cell gastric adenocarcinoma (b) and the proband’s unaffected normal gastric mucosa (c). Images obtained at 20×.
The proband of family 14 is a Hispanic female who had metastatic, poorly differentiated, diffuse signet-ring cell gastric cancer diagnosed at age 39 (Fig. 3a). IHC of a lymph node metastasis for α-E-catenin demonstrated decreased expression of α-E-catenin in the tumor compared with adjacent lymphoid tissue (Fig. 3b). The proband’s mother was diagnosed with gastric cancer at age 50 and died at age 51, and her maternal cousin was diagnosed with head and neck cancer at age 28. To our knowledge, no other members of family 14 have undergone CTNNA1 testing.
Finally, the proband of family 15 is a Caucasian female with grade 1, ER+/PR+/Her2- invasive ductal breast cancer diagnosed at age 53 (Fig. 4). This proband also had a history of a basal cell carcinoma at age 43. The family history is significant for her mother with pancreatic cancer diagnosed at age 94, a paternal cousin with brain cancer at age 35, and another paternal cousin with thyroid cancer at age 40. No other members of her family have undergone CTNNA1 testing.
DISCUSSION
MGPT includes analysis of well characterized, highly penetrant cancer risk genes, which are important for proper risk stratification for affected individuals and their families. However, the expansion of the number of genes available on MGPT has allowed for more widespread testing of “preliminary evidence genes” such as CTNNA1, which has been considered a potential diffuse gastric cancer susceptibility gene similar to CDH1. Testing for preliminary evidence genes has become increasingly important for better characterization of the associated clinical phenotypes. We report the results of germline CTNNA1 testing in a large cohort of individuals undergoing genetic testing at a commercial lab. We demonstrate that CTNNA1 LOF variants are detected at low rates in this large cohort, and that CTNNA1 LOF variants are found in individuals with personal and/or family histories of gastric and/or breast cancer.
In our cohort undergoing genetic testing, CTNNA1 LOF variants were found at a rate of 1 in 2912 individuals. This is a more frequent rate than that observed in the Genome Aggregation Database (gnomAD), where CTNNA1 LOF variants were observed in 1 in 17,682 individuals (8 CTNNA1 LOF variants of 141,456 individuals) (unpublished, Karczewski KJ, Francioli LC, Tiao G, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes, bioRxiv, 2019:531210). This higher rate of CTNNA1 LOF variants in our cohort suggests there may be enrichment in CTNNA1 LOF variants in a cohort ascertained for personal/family history of cancer when compared with the general population. However, CTNNA1 LOF variants were found less frequently than pathogenic/likely pathogenic CDH1 variants in the testing cohort, which were present in 106 of 151,425 individuals tested, for a rate of 1 in 1429 individuals.
CTNNA1 LOF variants were found previously in families meeting criteria for HDGC syndrome with a highly penetrant gastric cancer phenotype.7,8,9,10,11 However, two other reports of CTNNA1 LOF variants found no gastric cancer, including one individual identified with a CTNNA1 c.1183delC (p.Leu395Trpfs*10) variant in a large study of MGPT on unselected Chinese breast cancer patients,15 and a CTNNA1 c.2101G>T (p.Glu701*) variant identified in a study examining truncating protein variants in neurodevelopmental disorders.16 In our cohort, 12% of the individuals found to have a CTNNA1 LOF variant had a history of diffuse gastric cancer, while 21% of those found to have a CTNNA1 LOF variant had either a personal or family history of gastric cancer. Although there was enrichment for gastric cancer in CTNNA1 LOF variant carriers, the rate of gastric cancer observed in our cohort may appear surprisingly lower than expected given prior reports strongly associating CTNNA1 with gastric cancer.7,8,9,10,11 However, decreased penetrance estimates are seen in CDH1 carriers when analyzing unselected cohorts rather than highly penetrant families, thus decreasing the selection bias.17,18
CDH1 variants have also been associated with increased lobular breast cancer risk.19,20 In examining personal and family history of breast cancer in CTNNA1 LOF carriers, we found 67% of carriers had a history of breast cancer, while 83% of carriers had either a personal history of or a first-degree relative with breast cancer. This high percentage may be influenced by the population undergoing genetic testing, where over one-third of individuals tested had a personal history of breast cancer. However, despite the high number of individuals with a personal history of breast cancer in the testing cohort, there was still statistically significant enrichment for breast cancer in CTNNA1 LOF carriers. Given the limitations in data collection, it is unknown if the majority of breast cancer seen in CTNNA1 LOF families is predominantly ductal or lobular, which prevents determination of whether the observed breast cancer enrichment is specific for lobular and/or ductal breast cancers. However, while most individuals did not specify the breast cancer subtype, the majority of breast cancers where a subtype was reported were ductal. It is also worthwhile to note that although many CTNNA1 LOF carriers had a personal history of breast cancer, the prevalence of LOF variants among individuals with breast cancer was low at 0.04% (22 of 52,093).
Of additional importance is whether CTNNA1 LOF variants are directly involved in increasing gastric and/or breast cancer risk. CTNNA1 codes for α-E-catenin, which interacts with the cell–cell adhesion molecule E-cadherin, the protein product of CDH1.21 Prior studies demonstrated that α-E-catenin expression is lost in gastric cancers of individuals carrying CTNNA1 LOF variants, thus inferring a potential role in gastric carcinogenesis.7,8 We observed similar findings in our families with the CTNNA1 c.1351C>T nonsense variant, where the early-onset diffuse gastric cancers showed decreased α-E-catenin expression. This evidence points toward a potential association of CTNNA1 LOF variants with increased gastric cancer risk, especially for certain variants such as CTNNA1 c.1351C>T; however, further study of the biological mechanism of this potential association is needed. The site of truncation may play a role in mediating risk. In families 23–29 the truncations are located in the C-terminus and would not be expected to lead to nonsense-mediated decay, which is of potential interest as none of these individuals nor their first-degree family members were reported to have a history of gastric cancer.
A challenge of testing preliminary evidence genes is deciding when to designate LOF variants as pathogenic/likely pathogenic given insufficient evidence for a disease-causing role.22 Given the limited evidence, commercial labs may not call CTNNA1 LOF variants pathogenic/likely pathogenic, which is standard practice when the gene–disease association is uncertain, and instead all of these variants may be categorized as VUSs. Therefore it is incumbent on the ordering clinician to closely examine any CTNNA1 variant discovered in the context of other clinical and family history information to properly utilize these results for appropriate clinical care. It is likely that clinical guidelines for management of CTNNA1 LOF carriers, for example from the International Gastric Cancer Linkage Consortium (IGCLC), may help enable commercial labs to call these variants pathogenic/likely pathogenic. Further complicating matters with CTNNA1 is the rate of missense VUSs identified. Including the 1057 missense VUSs with the 52 LOF variants, 0.7% of the tested population was identified to have a CTNNA1 VUS.
The management of individuals with CTNNA1 LOF variants remains challenging. Currently, there are no guidelines or expert opinion panels that provide concrete clinical recommendations for CTNNA1 LOF carriers, and therefore, management of these individuals must be individualized. For gastric cancer risk, while a risk-reducing total gastrectomy for a CTNNA1 LOF variant was reported,8 given the preliminary evidence surrounding CTNNA1 as well as the unclear and possibly lower than expected gastric cancer risk, risk-reducing total gastrectomy will likely not be a reasonable choice for all individuals with CTNNA1 variants at this time. However, whether or not to pursue gastric cancer surveillance, in a similar manner to CDH1 carriers who defer gastrectomy, can be considered. While it has been reported that invasive lobular carcinoma is more common in CDH1 carriers,19,20 we do not know if this will also pertain to CTNNA1, as the majority of our cohort who reported a personal history of breast cancer did not specify pathology. Breast cancer management of these individuals will also need to be addressed as more data are generated; however, due to limited evidence, we do not suggest altering breast cancer surveillance in CTNNA1 carriers at this time, and therefore the recommendation for breast cancer screening should be individualized based on personal and family history of the disease, not the presence or absence of CTNNA1 LOF variants.
Given the uncertainties in management, it is important that individuals who undergo genetic testing for preliminary evidence genes, such as CTNNA1, have thorough pretest genetic counseling. Furthermore, CTNNA1 LOF carriers should be strongly encouraged to enroll in research registries to allow accurate collection of personal and family history, as well as collection of longitudinal outcomes, since these data will be critical for refining the cancer risk spectrum of CTNNA1.
There are several limitations in our study. First, other than the three CTNNA1 c.1351C>T families, personal and family history data were obtained from genetic testing requisition forms, and unable to be confirmed by direct review of the medical records. However, recent data have demonstrated that ordering clinicians typically do an accurate job of reporting personal and family history on requisition forms.23 Another limitation to our study is that the breast cancer type was provided in less than half of the reported breast cancers in the CTNNA1 LOF carriers. Additionally, individuals undergoing CTNNA1 requisitioning are subject to selection bias as these individuals primarily had a personal or family history of cancer. Therefore, it is possible that if CTNNA1 testing was performed in a completely unselected population, there may be a lower number of gastric and/or breast cancers in CTNNA1 families than we have reported. Finally, given the small number of CTNNA1 LOF variant carriers as well as the limited complete family history for the majority of these carriers, calculation of accurate cancer risk estimates was not feasible.
This study provides the first large-scale report of CTNNA1 LOF variants in individuals undergoing genetic testing for cancer risk assessment. We demonstrate that CTNNA1 LOF variants are detected on MGPT with a majority of the individuals with a LOF variant having breast cancer and a minority having gastric cancer. These data show that the gastric cancer risk associated with a CTNNA1 LOF variant may be lower than initially predicted; however, there may be specific variants, such as the CTNNA1 nonsense variant c.1351C>T, which may be a significant risk factor for early-onset diffuse gastric cancer. Management of individuals with CTNNA1 LOF variants remains challenging, and capturing affected individuals in prospective registries will be critical to better determine the true cancer risks associated with CTNNA1 LOF variants.
References
Easton DF, Pharoah PD, Antoniou AC, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372:2243–2257.
Slavin TP, Niell-Swiller M, Solomon I, et al. Clinical application of multigene panels: challenges of next-generation counseling and cancer risk management. Front Oncol. 2015;5:208.
American Cancer Society. Global cancer facts & figures. 4th edition. Atlanta: American Cancer Society; 2018.
Cislo M, Filip AA, Arnold Offerhaus GJ, et al. Distinct molecular subtypes of gastric cancer: from Lauren to molecular pathology. Oncotarget. 2018;9:19427–19442.
van der Post RS, Oliveira C, Guilford P, Carneiro F. Hereditary gastric cancer: what’s new? Update 2013-2018. Fam Cancer. 2019;18:363–367.
van der Post RS, Vogelaar IP, Carneiro F, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Genet. 2015;52:361–374.
Majewski IJ, Kluijt I, Cats A, et al. An alpha-E-catenin (CTNNA1) mutation in hereditary diffuse gastric cancer. J Pathol. 2013;229:621–629.
Benusiglio PR, Colas C, Guillerm E, et al. Clinical implications of CTNNA1 germline mutations in asymptomatic carriers. Gastric Cancer. 2019;22:899–903.
Weren RDA, van der Post RS, Vogelaar IP, et al. Role of germline aberrations affecting CTNNA1, MAP3K6 and MYD88 in gastric cancer susceptibility. J Med Genet. 2018;55:669–674.
van der Post RS, van Dieren J, Grelack A, et al. Outcomes of screening gastroscopy in first-degree relatives of patients fulfilling hereditary diffuse gastric cancer criteria. Gastrointest Endosc. 2018;87:397–404 e392.
Hansford S, Kaurah P, Li-Chang H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1:23–32.
Kumar S, Long JM, Ginsberg GG, Katona BW. The role of endoscopy in the management of hereditary diffuse gastric cancer syndrome. World J Gastroenterol. 2019;25:2878–2886.
Lincoln SE, Kobayashi Y, Anderson MJ, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17:533–544.
Nykamp K, Anderson M, Powers M, et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19:1105–1117.
Sun J, Meng H, Yao L, et al. Germline mutations in cancer susceptibility genes in a large series of unselected breast cancer patients. Clin Cancer Res. 2017;23:6113–6119.
Kosmicki JA, Samocha KE, Howrigan DP, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49:504–510.
Roberts ME, Ranola JMO, Marshall ML, et al. Comparison of CDH1 penetrance estimates in clinically ascertained families vs families ascertained for multiple gastric cancers. JAMA Oncol. 2019;5:1325–1331.
Xicola RM, Li S, Rodriguez N, et al. Clinical features and cancer risk in families with pathogenic CDH1 variants irrespective of clinical criteria. J Med Genet. 2019;56:838–843.
Chikman B, Davidson T, Kais H, et al. Is there an association between invasive lobular carcinoma of the breast and a family history of gastric cancer? Fam Cancer. 2016;15:41–47.
Benusiglio PR, Malka D, Rouleau E, et al. CDH1 germline mutations and the hereditary diffuse gastric and lobular breast cancer syndrome: a multicentre study. J Med Genet. 2013;50:486–489.
Vite A, Li J, Radice GL. New functions for alpha-catenins in health and disease: from cancer to heart regeneration. Cell Tissue Res. 2015;360:773–783.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424.
LaDuca H, McFarland R, Gutierrez S, et al. Quality of clinician-reported cancer history when ordering genetic testing. JCO Clin Cancer Inform. 2018;2:1–11.
Acknowledgements
We would like to acknowledge Shan Yang from Invitae for all of her assistance with data acquisition and processing. This work was funded by National Institutes of Health (NIH)/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants K08DK106489 and R03DK120946 (B.W.K.) and The Lustgarten Family Colon Cancer Research Fund (A.K.R., B.W.K.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The following conflicts of interest are reported: B.N.: advisory board (Invitae); S.T.M., D.P.-A., M.J.A.: employment (Invitae), Shareholder (Invitae); S.M.D.: honoraria (AstraZeneca, Clovis, Bristol-Myers Squibb); B.W.K.: consulting (Exact Sciences), travel (Janssen). The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Clark, D.F., Michalski, S.T., Tondon, R. et al. Loss-of-function variants in CTNNA1 detected on multigene panel testing in individuals with gastric or breast cancer. Genet Med 22, 840–846 (2020). https://doi.org/10.1038/s41436-020-0753-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0753-1
Keywords
This article is cited by
-
Current advances in understanding the molecular profile of hereditary diffuse gastric cancer and its clinical implications
Journal of Experimental & Clinical Cancer Research (2023)
-
Combining clinical and molecular characterization of CDH1: a multidisciplinary approach to reclassification of a splicing variant
Familial Cancer (2023)
-
Mammary Lobular Carcinoma-Like Salivary Gland Carcinoma: Report of a Rare Case
Head and Neck Pathology (2022)
-
Genetic Gastric Cancer Risk Syndromes
Current Treatment Options in Gastroenterology (2020)