By Andy Flick, Evolutionary Studies Initiative scientific coordinator
Jacob Steenwyk, a graduate student in biological sciences, and collaborators at Vanderbilt, Tel Aviv University and University of Wisconsin-Madison measured the coevolution of pairs of genes shared across budding yeasts to identify the genes that participate in the same cellular or molecular functions.
The results of this work could fundamentally change how we identify genes with similar functions.
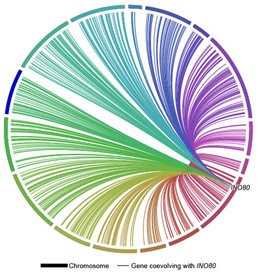
“In this project, we examined nearly 3 million pairs of genes and identified instances where pairs of genes had strong evidence of coevolution,” Steenwyk said. “This allowed us to draw a network diagram that reflected cellular and genomic structure and function.”
This project shows that genes’ evolutionary histories yield insights similar to those of genetic studies on model organisms. This is a hugely important insight because evolutionary analyses are often far less challenging and require fewer resources than genetic studies.
The work was made possible by PhyKIT, a new software co-developed by Steenwyk, and genomic data from hundreds of genomes. PhyKIT is a novel way to use the evolutionary histories of genes to inform our understanding of how proteins interact inside the cell. The target group of organisms used were budding yeasts, a group of microscopic fungi that have a plethora of evolutionary and genetic data available.
“This manuscript is the first major project that heavily leaned on the software I developed,” Steenwyk said. “In particular, I used PhyKIT, a broadly applicable toolkit for analyzing and processing multiple sequence alignments and phylogenies.”
While Steenwyk remains humble—noting that this area is something of “novel terrain”—his software products have been downloaded more than a hundred thousand times. “Jacob saw a need among evolutionary biologists for user-friendly, robust software. He not only rose to this challenge by developing numerous pieces of software but integrated software development into his approach for asking how the evolution of genes can inform us about their function,” Rokas said.
Another interesting result is that gene function, rather than gene location within a chromosome, drives the coevolution of genes. According to the authors, genomes may best be viewed as extensively linked groups of genes. “Our project sets the stage for examining gene coevolution networks in emerging model organisms, lineages with less functional data or lineages that are not yet genetically tractable,” Steenwyk said.
Steenwyk recently received the 2022 Harold M. Weintraub Graduate Student Award, an international award for outstanding achievement in graduate work in biological sciences and defended his Ph.D. in the lab of Antonis Rokas, Cornelius Vanderbilt Chair in Biological Sciences and director of the Evolutionary Studies Initiative.

The article “An orthologous gene coevolution network provides insight into eukaryotic cellular and genomic structure and function,” was published in the journal Science Advances, on May 4.
Steenwyk and Rokas were supported by the Howard Hughes Medical Institute through the James H. Gilliam Fellowships for Advanced Study program. The Rokas lab is also supported by funds from the Burroughs Wellcome Fund, the National Science Foundation grants DEB-1442113 and DEB-2110404 and the National Institutes of Health/National Institute of Allergy and Infectious Diseases grants R56 AI146096 and R01 AI153356.