Self-Amplifying RNA Approach for Protein Replacement Therapy
Abstract
:1. Introduction
2. Clinical Trials of mRNA Approach for Protein Replacement Therapy
3. Major Differences between saRNA & mRNA Technologies
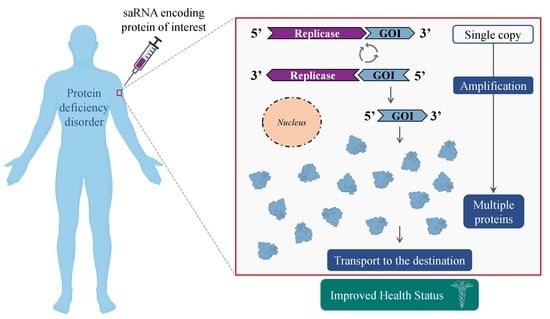
4. saRNA—Mechanism of Action
5. Potential Application of saRNA for Non-Infectious Health Disorders
6. saRNA for Single-Gene Disorders—Special Focus on AATD
7. saRNA for Cancer Immunotherapy
8. Delivery Systems for saRNA Therapeutics
9. Challenges
10. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Damase, T.R.; Sukhovershin, R.; Boada, C.; Taraballi, F.; Pettigrew, R.I.; Cooke, J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 628137. [Google Scholar] [CrossRef] [PubMed]
- Blakney, A. The next generation of RNA vaccines: Self-amplifying RNA. Biochemist 2021, 43, 14–17. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Blakney, A.K.; Ip, S.; Geall, A.J. An update on self-amplifying mRNA vaccine development. Vaccines 2021, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Bloom, K.; van den Berg, F.; Arbuthnot, P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2020, 28, 117–129. [Google Scholar] [CrossRef]
- Vogel, A.B.; Lambert, L.; Kinnear, E.; Busse, D.; Erbar, S.; Reuter, K.C.; Wicke, L.; Perkovic, M.; Beissert, T.; Haas, H.; et al. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol. Ther. 2018, 26, 446–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magadum, A.; Kaur, K.; Zangi, L. mRNA-Based Protein Replacement Therapy for the Heart. Mol. Ther. 2019, 27, 785–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karadagi, A.; Cavedon, A.G.; Zemack, H.; Nowak, G.; Eybye, M.E.; Zhu, X.; Guadagnin, E.; White, R.A.; Rice, L.M.; Frassetto, A.L.; et al. Systemic modified messenger RNA for replacement therapy in alpha 1-antitrypsin deficiency. Sci. Rep. 2020, 10, 7052. [Google Scholar] [CrossRef]
- Fessel, J. A vaccine to prevent initial loss of cognition and eventual Alzheimer’s disease in elderly persons. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2021, 7, e12126. [Google Scholar] [CrossRef]
- Borah, P.; Deb, P.K.; Al-Shar’i, N.A.; Dahabiyeh, L.A.; Venugopala, K.N.; Singh, V.; Shinu, P.; Hussain, S.; Deka, S.; Chandrasekaran, B.; et al. Perspectives on RNA Vaccine Candidates for COVID-19. Front. Mol. Biosci. 2021, 8, 635245. [Google Scholar] [CrossRef]
- Spencer, A.J.; Mckay, P.F.; Belij-rammerstorfer, S.; Ulaszewska, M.; Bissett, C.D.; Hu, K.; Samnuan, K.; Blakney, A.K.; Wright, D.; Sharpe, H.R.; et al. Heterologous vaccination regimens with self-amplifying RNA and adenoviral COVID vaccines induce robust immune responses in mice. Nat. Commun. 2021, 12, 2893. [Google Scholar] [CrossRef] [PubMed]
- De Alwis, R.; Gan, E.S.; Chen, S.; Leong, Y.S.; Tan, H.C.; Zhang, S.L.; Yau, C.; Low, J.G.H.; Kalimuddin, S.; Matsuda, D.; et al. A single dose of self-transcribing and replicating RNA-based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. Mol. Ther. 2021, 29, 1970–1983. [Google Scholar] [CrossRef] [PubMed]
- Fenton, C.; Lamb, Y.N. COVID-19: State of the Vaccination. Drugs Ther. Perspect. 2021, 37, 508–518. [Google Scholar] [CrossRef]
- Papukashvili, D.; Rcheulishvili, N.; Liu, C.; Xie, F.; Tyagi, D.; He, Y.; Wang, P.G. Perspectives on miRNAs Targeting DKK1 for Developing Hair Regeneration Therapy. Cells 2021, 10, 2957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Maruggi, G.; Shan, H.; Li, J. Advances in mRNA Vaccines for Infectious Diseases. Front. Immunol. 2019, 10, 594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollock, K.M.; Cheeseman, H.M.; Szubert, A.J.; Libri, V.; Boffito, M.; Owen, D.; Bern, H.; O’Hara, J.; McFarlane, L.R.; Lemm, N.M.; et al. Safety and immunogenicity of a self-amplifying RNA vaccine against COVID-19: COVAC1, a phase I, dose-ranging trial. eClinicalMedicine 2022, 44, 101262. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, K. Self-Replicating RNA Viruses for Vaccine Development against Infectious Diseases and Cancer. Vaccines 2021, 9, 1187. [Google Scholar] [CrossRef]
- Li, Y.; Su, Z.; Zhao, W.; Zhang, X.; Momin, N.; Zhang, C.; Wittrup, K.D.; Dong, Y.; Irvine, D.J.; Weiss, R. Multifunctional oncolytic nanoparticles deliver self-replicating IL-12 RNA to eliminate established tumors and prime systemic immunity. Nat. Cancer 2020, 1, 882–893. [Google Scholar] [CrossRef]
- Li, Y.; Teague, B.; Zhang, Y.; Su, Z.; Porter, E.; Dobosh, B.; Wagner, T.; Irvine, D.J.; Weiss, R. In vitro evolution of enhanced RNA replicons for immunotherapy. Sci. Rep. 2019, 9, 6932. [Google Scholar] [CrossRef] [Green Version]
- Miao, L.; Li, L.; Huang, Y.; Delcassian, D.; Chahal, J.; Han, J.; Shi, Y.; Sadtler, K.; Gao, W.; Lin, J.; et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 2019, 37, 1174–1185. [Google Scholar] [CrossRef]
- Sun, P.; Hernandez-Guillamón, M.; Campos-Martorell, M.; Simats, A.; Montaner, J.; Unzeta, M.; Solé, M. Simvastatin blocks soluble SSAO/VAP-1 release in experimental models of cerebral ischemia: Possible benefits for stroke-induced inflammation control. Biochim. Biophys. Acta—Mol. Basis Dis. 2018, 1864, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, L.; Almutairi, M.M.; Hotez, P.J.; Pollet, J. Enlisting the mRNA Vaccine Platform to Combat Parasitic Infections. Vaccines 2019, 7, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, A.; Kormann, M.; Rosenecker, J.; Rudolph, C. Current prospects for mRNA gene delivery. Eur. J. Pharm. Biopharm. 2009, 71, 484–489. [Google Scholar] [CrossRef]
- Xu, S.; Yang, K.; Li, R.; Zhang, L. mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection. Int. J. Mol. Sci. 2020, 21, 6582. [Google Scholar] [CrossRef] [PubMed]
- Kormann, M.S.D.; Hasenpusch, G.; Aneja, M.K.; Nica, G.; Flemmer, A.W.; Herber-jonat, S.; Huppmann, M.; Mays, L.E.; Illenyi, M.; Schams, A.; et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011, 29, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Michel, T.; Wendel, H.; Krajewski, S. Next-Generation Therapeutics: mRNA as a Novel Therapeutic Option for Single-Gene Disorders. In Modern Tools for Genetic Engineering; Kormann, M., Ed.; IntechOpen: London, UK, 2016; pp. 3–20. [Google Scholar]
- Maruggi, G.; Ulmer, J.B.; Rappuoli, R.; Yu, D. Self-amplifying mRNA-Based Vaccine Technology and Its Mode of Action. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar] [CrossRef]
- Sandbrink, J.B.; Shattock, R.J. RNA Vaccines: A Suitable Platform for Tackling Emerging Pandemics? Front. Immunol. 2020, 11, 608460. [Google Scholar] [CrossRef]
- Fros, J.J.; Pijlman, G.P. Alphavirus infection: Host cell shut-off and inhibition of antiviral responses. Viruses 2016, 8, 166. [Google Scholar] [CrossRef] [Green Version]
- Götte, B.; Liu, L.; Mcinerney, G.M. The Enigmatic Alphavirus Non-Structural Protein 3 (nsP3) Revealing Its Secrets at Last. Viruses 2018, 10, 105. [Google Scholar] [CrossRef] [Green Version]
- Ballesteros-Briones, M.C.; Silva-Pilipich, N.; Herrador-Canete, G.; Vanrell, L.; Smerdou, C. A new generation of vaccines based on alphavirus self-amplifying RNA. Curr. Opin. Virol. 2020, 44, 145–153. [Google Scholar] [CrossRef]
- Blakney, A.K.; McKay, P.F.; Shattock, R.J. Structural Components for Amplification of Positive and Negative Strand VEEV Splitzicons. Front. Mol. Biosci. 2018, 5, 71. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Eygeris, Y.; Gupta, M.; Sahay, G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021, 170, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.A.; Kommareddy, S.; Maione, D.; Uematsu, Y.; Giovani, C.; Scorza, F.B.; Otten, G.R.; Yu, D.; Mandl, C.W.; Mason, P.W.; et al. Self-Amplifying mRNA Vaccines; Elsevier Ltd.: Amsterdam, The Netherlands, 2015. [Google Scholar] [CrossRef]
- Fuller, D.H.; Berglund, P. Amplifying RNA Vaccine Development. N. Engl. J. Med. 2020, 382, 2469–2471. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Atasheva, S.; McAuley, A.J.; Plante, J.A.; Frolova, E.I.; Beasley, D.W.C.; Frolov, I. Enhancement of protein expression by alphavirus replicons by designing self-replicating subgenomic RNAs. Proc. Natl. Acad. Sci. USA 2014, 111, 10708–10713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pepini, T.; Pulichino, A.-M.; Carsillo, T.; Carlson, A.L.; Sari-Sarraf, F.; Ramsauer, K.; Debasitis, J.C.; Maruggi, G.; Otten, G.R.; Geall, A.J.; et al. Induction of an IFN-Mediated Antiviral Response by a Self-Amplifying RNA Vaccine: Implications for Vaccine Design. J. Immunol. 2017, 198, 4012–4024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minnaert, A.; Vanluchene, H.; Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Raemdonck, K.; Sanders, N.N.; Remaut, K. Strategies for controlling the innate immune activity of conventional and self-amplifying mRNA therapeutics: Getting the message across. Adv. Drug Deliv. Rev. 2021, 176, 113900. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Am. Soc. Gene Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Blakney, A.K.; Mckay, P.F.; Bouton, C.R.; Hu, K.; Samnuan, K.; Shattock, R.J. Innate Inhibiting Proteins Enhance Expression and Immunogenicity of Self-Amplifying RNA. Mol. Ther. 2021, 29, 1174–1185. [Google Scholar] [CrossRef]
- Gómez-aguado, I.; Rodríguez-castejón, J.; Vicente-pascual, M.; Rodríguez-, A.; Solinís, M.Á.; Pozo-rodríguez, A. Nanomedicines to Deliver mRNA: State of the Art and Future Perspectives. Nanomaterials 2020, 10, 364. [Google Scholar] [CrossRef]
- Leon, B.M.; Maddox, T.M. Diabetes and cardiovascular disease: Epidemiology, biological mechanisms, treatment recommendations and future research. World J. Diabetes 2015, 6, 1246–1258. [Google Scholar] [CrossRef]
- Sapra, A.; Bhandari, P. Diabetes Mellitus. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021; pp. 1–15. [Google Scholar]
- Papukashvili, D.; Rcheulishvili, N.; Deng, Y. Beneficial Impact of Semicarbazide-Sensitive Amine Oxidase Inhibition on the Potential Cytotoxicity of Creatine Supplementation in Type 2 Diabetes Mellitus. Molecules 2020, 25, 2029. [Google Scholar] [CrossRef]
- Leutner, M.; Haug, N.; Bellach, L.; Dervic, E.; Kautzky, A.; Klimek, P.; Kautzky-willer, A. Risk of Typical Diabetes-Associated Complications in Different Clusters of Diabetic Patients: Analysis of Nine Risk Factors. J. Pers. Med. 2021, 11, 328. [Google Scholar] [CrossRef]
- Christ-Crain, M.; Winzeler, B.; Refardt, J. Diagnosis and management of diabetes insipidus for the internist: An update. J. Intern. Med. 2021, 290, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Moeller, H.B.; Rittig, S.; Fenton, R.A. Nephrogenic Diabetes Insipidus: Essential Insights into the Molecular Background and Potential Therapies for Treatment. Endocr. Rev. 2013, 34, 278–301. [Google Scholar] [CrossRef] [Green Version]
- Kalra, S.; Zargar, A.H.; Jain, S.M.; Sethi, B.; Chowdhury, S. Review Article Diabetes insipidus: The other diabetes. Indian J. Endocrinol. Metab. 2016, 20, 9–21. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Schneller, J.L.; Guey, L.T.; Charles, P.; Martini, P.G.V.; An, D.; Schneller, J.L.; Frassetto, A.; Liang, S.; Zhu, X.; et al. Systemic Messenger RNA Therapy as a Treatment for Methylmalonic Acidemia Systemic Messenger RNA Therapy as a Treatment for Methylmalonic Acidemia. Cell Rep. 2017, 21, 3548–3558. [Google Scholar] [CrossRef] [Green Version]
- Kmiec, D.; Kirchhoff, F. Monkeypox: A New Threat? Int. J. Mol. Sci. 2022, 23, 7866. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Langer, R.; Wechsler, M.E.; Peppas, N.A. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jia, C.; Yang, J. Designing Nanoparticle-based Drug Delivery Systems for Precision Medicine. Int. J. Med. Sci. 2021, 18, 2943–2949. [Google Scholar] [CrossRef]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef] [Green Version]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; Dipiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Pardi, N.; Tuyishime, S.; Muramatsu, H.; Kariko, K.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Weissman, D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release Off. J. Control. Release Soc. 2010, 217, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Pardi, N.; Parkhouse, K.; Kirkpatrick, E.; McMahon, M.; Zost, S.J.; Mui, B.L.; Tam, Y.K.; Karikó, K.; Barbosa, C.J.; Madden, T.D.; et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018, 9, 3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samnuan, K.; Blakney, A.K.; Mckay, P.F.; Shattock, R.J. Design-of-experiments in vitro transcription yield optimization of self-amplifying RNA [version 1; peer review: 1 approved with reservations]. F1000Research 2022, 11, 333. [Google Scholar] [CrossRef]
- Maruggi, G.; Mallett, C.P.; Westerbeck, J.W.; Chen, T.; Lofano, G.; Friedrich, K.; Qu, L.; Sun, J.T.; Mcauliffe, J.; Kanitkar, A.; et al. A self-amplifying mRNA SARS-CoV-2 vaccine candidate induces safe and robust protective immunity in preclinical models. Mol. Ther. 2022, 30, 1897–1912. [Google Scholar] [CrossRef] [PubMed]
- Geall, A.J.; Verma, A.; Otten, G.R.; Shaw, C.A.; Hekele, A.; Banerjee, K.; Cu, Y.; Valiante, N.M.; Dormitzer, P.R.; Barnett, S.W.; et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA 2012, 109, 14604–14609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, G.; Anderluzzi, G.; Tandrup, S.; Woods, S.; Gallorini, S.; Brazzoli, M.; Giusti, F.; Ferlenghi, I.; Johnson, R.N.; Roberts, C.W.; et al. Delivery of self-amplifying mRNA vaccines by cationic lipid nanoparticles: The impact of cationic lipid selection. J. Control. Release 2020, 325, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, H.; Moorthie, S.; Petrou, M.; Hamamy, H.; Povey, S.; Bittles, A.; Gibbons, S.; Darlison, M.; Modell, B.; Disorders, C.; et al. Rare single gene disorders: Estimating baseline prevalence and outcomes worldwide. J. Community Genet. 2018, 9, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchini, M.; Mannucci, P.M. Haemophilia B is clinically less severe than haemophilia A: Further evidence. Blood Transfus. 2018, 16, 121–122. [Google Scholar] [CrossRef]
- Chapman, K.R.; Chorostowska-Wynimko, J.; Koczulla, A.R.; Ferrarotti, I.; McElvaney, N.G. Alpha 1 antitrypsin to treat lung disease in alpha 1 antitrypsin deficiency: Recent developments and clinical implications. Int. J. Chron. Obstruct. Pulmon. Dis. 2018, 13, 419–432. [Google Scholar] [CrossRef] [Green Version]
- Gershon, A.S.; Thiruchelvam, D.; Chapman, K.R.; Aaron, S.D.; Stanbrook, M.B.; Bourbeau, J.; Tan, W.; To, T.; Respiratory, C. Health Services Burden of Undiagnosed and Overdiagnosed COPD. Chest 2018, 153, 1336–1346. [Google Scholar] [CrossRef] [PubMed]
- Adeloye, D.; Song, P.; Zhu, Y.; Campbell, P.H.; Sheikh, P.A.; Rudan, P.I.; Respire, N.; Respiratory, G.; Unit, H. Global, regional, and national prevalence of, and risk factors for, chronic obstructive pulmonary disease (COPD) in 2019: A systematic review and modelling analysis. Lancet Respir. 2022, 10, 447–458. [Google Scholar] [CrossRef]
- Tejwani, V.; Stoller, J.K. The spectrum of clinical sequelae associated with alpha-1 antitrypsin deficiency. Ther. Adv. Chronic Dis. 2021, 12, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Laurell, C.B.; Eriksson, S. The electrophoretic α1-globulin pattern of serum in α1-antitrypsin deficiency. COPD J. Chronic Obstr. Pulm. Dis. 2013, 10, 3–8. [Google Scholar] [CrossRef]
- Stoller, J.K.; Hupertz, V.; Aboussouan, L.S. Alpha-1 antitrypsin deficiency. In Gene Reviews; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2020; pp. 34–35. ISBN 9781975126841. [Google Scholar]
- Barriga, V.; Kuol, N.; Nurgali, K.; Apostolopoulos, V. The complex interaction between the tumor micro-environment and immune checkpoints in breast cancer. Cancers 2019, 11, 1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apostolopoulos, V. Cancer vaccines: Research and applications. Cancers 2019, 11, 1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neek, M.; Kim, T., II; Wang, S.W. Protein-based nanoparticles in cancer vaccine development. Nanomed. Nanotechnol. Biol. Med. 2019, 15, 164–174. [Google Scholar] [CrossRef]
- Morse, M.A.; Gwin, W.R., III; Mitchell, A.D. Vaccine Therapies for Cancer: Then and Now; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; Volume 16, ISBN 1152302000. [Google Scholar]
- Wang, Y.; Zhang, Z.; Luo, J.; Han, X.; Wei, Y. mRNA vaccine: A potential therapeutic strategy. Mol. Cancer 2021, 20, 33. [Google Scholar] [CrossRef]
- Heine, A.; Juranek, S.; Brossart, P. Clinical and immunological effects of mRNA vaccines in malignant diseases. Mol. Cancer 2021, 20, 52. [Google Scholar] [CrossRef]
- Moderna Moderna Research Product Pipeline. Available online: https://www.modernatx.com/research/product-pipeline (accessed on 19 October 2022).
- Blakney, A.K.; Zhu, Y.; McKay, P.F.; Bouton, C.R.; Yeow, J.; Tang, J.; Hu, K.; Samnuan, K.; Grigsby, C.L.; Shattock, R.J.; et al. Big Is Beautiful: Enhanced saRNA Delivery and Immunogenicity by a Higher Molecular Weight, Bioreducible, Cationic Polymer. ACS Nano 2020, 14, 5711–5727. [Google Scholar] [CrossRef] [Green Version]
- Blakney, A.K.; Mckay, P.F.; Hu, K.; Samnuan, K.; Jain, N.; Brown, A.; Thomas, A.; Rogers, P.; Polra, K.; Sallah, H.; et al. Polymeric and lipid nanoparticles for delivery of self-amplifying RNA vaccines. J. Control. Release 2021, 338, 201–210. [Google Scholar] [CrossRef]
- McKay, P.F.; Hu, K.; Blakney, A.K.; Samnuan, K.; Brown, J.C.; Penn, R.; Zhou, J.; Bouton, C.R.; Rogers, P.; Polra, K.; et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020, 11, 3523. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, K. Nanoparticle-based delivery of self-amplifying RNA. Gene Ther. 2020, 27, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Kon, E.; Elia, U.; Peer, D. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID-19. The COVID-19 resource centre is hosted on Elsevier Connect, the company’s public news and information. Curr. Opin. Biotechnol. 2020, 73, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef] [Green Version]
- Alfagih, I.M.; Aldosari, B.; Alquadeib, B.; Almurshedi, A. Nanoparticles as Adjuvants and Nanodelivery Systems for mRNA-Based Vaccines. Pharmaceutics 2021, 13, 45. [Google Scholar] [CrossRef]
- Arteta, M.Y.; Kjellman, T.; Bartesaghi, S.; Wallin, S.; Wu, X.; Kvist, A.J.; Dabkowska, A.; Székely, N.; Radulescu, A.; Bergenholtz, J.; et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. USA 2018, 115, E3351–E3360. [Google Scholar] [CrossRef] [Green Version]
- Sabnis, S.; Kumarasinghe, E.S.; Salerno, T.; Mihai, C.; Ketova, T.; Senn, J.J.; Lynn, A.; Bulychev, A.; McFadyen, I.; Chan, J.; et al. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther. 2018, 26, 1509–1519. [Google Scholar] [CrossRef]
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q. Lipid Nanoparticles from Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. [Google Scholar] [CrossRef]
- Blakney, A.K.; McKay, P.F.; Yus, B.I.; Aldon, Y.; Shattock, R.J. Inside out: Optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019, 26, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Blakney, A.K.; McKay, P.F.; Yus, B.I.; Hunter, J.E.; Dex, E.A.; Shattock, R.J. The Skin You Are In: Design-of-Experiments Optimization of Lipid Nanoparticle Self- Amplifying RNA Formulations in Human Skin Explants. ACS Nano 2019, 13, 5920–5930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, E.; Liu, X.; Li, M.; Zhang, Z.; Song, L.; Zhu, B.; Wu, X.; Liu, J.; Zhao, D.; Li, Y. Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target. Ther. 2022, 7, 94. [Google Scholar] [CrossRef]
- Rodríguez-gascón, A.; Pozo-rodríguez, A. Development of nucleic acid vaccines: Use of self-amplifying RNA in lipid nanoparticles. Int. J. Nanomed. 2014, 4, 1833–1843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hekele, A.; Bertholet, S.; Archer, J.; Gibson, D.G.; Palladino, G.; Brito, L.A.; Otten, G.R.; Brazzoli, M.; Buccato, S.; Bonci, A.; et al. Rapidly produced SAM H vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013, 2, e52. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Zheng, L.; Hu, Y.; Liu, S.; Wang, Y.; Xiong, Z. Induction of Protective Immunity against Toxoplasma gondii in Mice by Nucleoside Triphosphate Self-amplifying RNA Vaccine Encapsulated in Lipid Nanoparticle (LNP). Front. Microbiol. 2017, 8, 605. [Google Scholar] [CrossRef] [Green Version]
- Lazzaro, S.; Giovani, C.; Mangiavacchi, S.; Magini, D.; Baudner, B.; Geall, A.J.; De Gregorio, E.; Oro, U.D.; Buonsanti, C. CD8 T-cell priming upon mRNA vaccination is restricted to bone-marrow-derived antigen-presenting cells and may involve antigen transfer from myocytes. Immunology 2015, 146, 312–326. [Google Scholar] [CrossRef] [Green Version]
- Magini, D.; Giovani, C.; Mangiavacchi, S.; Maccari, S. Self-Amplifying mRNA Vaccines Expressing Multiple Conserved Influenza Antigens Confer Protection against Homologous and Heterosubtypic Viral Challenge. PLoS ONE 2016, 11, e0161193. [Google Scholar] [CrossRef] [Green Version]
- Melo, M.; Porter, E.; Zhang, Y.; Silva, M.; Li, N.; Dobosh, B.; Liguori, A.; Skog, P.; Landais, E.; Menis, S.; et al. Immunogenicity of RNA Replicons Encoding HIV Env Immunogens Designed for Self-Assembly into Nanoparticles. Mol. Ther. 2019, 27, 2080–2090. [Google Scholar] [CrossRef]
- Goswami, R.; Chatzikleanthous, D.; Lou, G.; Giusti, F.; Bonci, A.; Taccone, M.; Brazzoli, M.; Gallorini, S.; Ferlenghi, I.; Berti, F.; et al. Mannosylation of LNP Results in Improved Potency for Self-Amplifying RNA (SAM) Vaccines. ACS Infect. Dis. 2019, 5, 1546–1558. [Google Scholar] [CrossRef]
- Flemming, A. Self-amplifying RNA in lipid nanoparticles: A next-generation vaccine? Nat. Rev. Drug Discov. 2012, 11, 748–749. [Google Scholar] [CrossRef]
- Gerhardt, A.; Voigt, E.; Archer, M.; Reed, S.; Larson, E.; Van Hoeven, N.; Kramer, R.; Fox, C.; Casper, C. A flexible, thermostable nanostructured lipid carrier platform for RNA vaccine delivery. Mol. Ther. Methods Clin. Dev. 2022, 25, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.A.; Chan, M.; Shaw, C.A.; Hekele, A.; Carsillo, T.; Schaefer, M.; Archer, J.; Seubert, A.; Otten, G.R.; Beard, C.W.; et al. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014, 22, 2118–2129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brazzoli, M.; Magini, D.; Bonci, A.; Buccato, S.; Giovani, C.; Kratzer, R.; Zurli, V.; Mangiavacchi, S.; Casini, D.; Brito, L.M.; et al. Induction of Broad-Based Immunity and Protective Efficacy by Self-amplifying mRNA Vaccines Encoding Influenza Virus Hemagglutinin. J. Virol. 2016, 90, 332–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruggi, G.; Chiarot, E.; Giovani, C.; Buccato, S.; Bonacci, S.; Frigimelica, E.; Margarit, I.; Geall, A.; Bensi, G.; Maione, D. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine 2017, 35, 361–368. [Google Scholar] [CrossRef]
- Samsa, M.M.; Dupuy, L.C.; Beard, C.W.; Six, C.M.; Schmaljohn, C.S.; Mason, P.W.; Geall, A.J.; Ulmer, J.B.; Yu, D. Self-Amplifying RNA Vaccines for Venezuelan Equine Encephalitis Virus Induce Robust Protective Immunogenicity in Mice. Mol. Ther. 2019, 27, 850–865. [Google Scholar] [CrossRef] [Green Version]
- Stokes, A.; Pion, J.; Binazon, O.; Laffont, B.; Bigras, M.; Dubois, G.; Blouin, K.; Young, J.K.; Ringenberg, M.A.; Ben Abdeljelil, N.; et al. Nonclinical safety assessment of repeated administration and biodistribution of a novel rabies self-amplifying mRNA vaccine in rats: Toxicity and biodistribution of rabies SAM vaccine. Regul. Toxicol. Pharmacol. 2020, 113, 104648. [Google Scholar] [CrossRef]
- Bogers, W.M.; Oostermeijer, H.; Mooij, P.; Koopman, G.; Verschoor, E.J.; Davis, D.; Ulmer, J.B.; Brito, L.A.; Cu, Y.; Banerjee, K.; et al. Potent immune responses in rhesus macaques induced by nonviral delivery of a self-amplifying RNA vaccine expressing HIV type 1 envelope with a cationic nanoemulsion. J. Infect. Dis. 2015, 211, 947–955. [Google Scholar] [CrossRef] [Green Version]
- Anderluzzi, G.; Lou, G.; Gallorini, S.; Brazzoli, M.; Johnson, R.; O’hagan, D.T.; Baudner, B.C.; Perrie, Y. Investigating the impact of delivery system design on the efficacy of self-amplifying RNA vaccines. Vaccines 2020, 8, 212. [Google Scholar] [CrossRef]
- Manara, C.; Brazzoli, M.; Piccioli, D.; Taccone, M.; Oro, U.D.; Maione, D.; Frigimelica, E. Co-administration of GM-CSF expressing RNA is a powerful tool to enhance potency of SAM-based vaccines. Vaccine 2019, 37, 4204–4213. [Google Scholar] [CrossRef]
- Zhou, X.; Berglund, P.; Rhodes, G.; Parker, S.E.; Jondal, M.; Liljestrom, P. Self-replicating Semliki Forest virus RNA as recombinant vaccine. Vaccine 1994, 12, 1510–1514. [Google Scholar] [CrossRef]
- Fleeton, M.N.; Chen, M.; Berglund, P.; Rhodes, G.; Parker, S.E.; Murphy, M.; Atkins, G.J.; Liljestro, P. Self-Replicative RNA Vaccines Elicit Protection against Influenza A Virus, Respiratory Syncytial Virus, and a Tickborne Encephalitis Virus. J. Infect. Dis. 2001, 183, 1395–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beissert, T.; Perkovic, M.; Vogel, A.; Erbar, S.; Walzer, K.C.; Hempel, T.; Brill, S.; Haefner, E.; Becker, R.; Türeci, Ö.; et al. A Trans-amplifying RNA Vaccine Strategy for Induction of Potent Protective Immunity. Mol. Ther. 2020, 28, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Portela Catani, J.P.; Mc Cafferty, S.; Couck, L.; Van Den Broeck, W.; Gorlé, N.; Vandenbroucke, R.E.; Devriendt, B.; Ulbert, S.; Cnops, L.; et al. Immunogenicity and Protection Efficacy of a Naked Self-Replicating mRNA-Based Zika Virus Vaccine. Vaccines 2019, 7, 96. [Google Scholar] [CrossRef] [Green Version]
- Cu, Y.; Broderick, K.E.; Banerjee, K.; Hickman, J.; Otten, G.; Barnett, S.; Kichaev, G.; Sardesai, N.Y.; Ulmer, J.B.; Geall, A. Enhanced Delivery and Potency of Self-Amplifying mRNA Vaccines by Electroporation in Situ. Vaccines 2013, 1, 367–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mccullough, K.C.; Bassi, I.; Milona, P.; Suter, R.; Thomann-harwood, L.; Englezou, P.; Démoulins, T.; Ruggli, N. Self-replicating Replicon-RNA Delivery to Dendritic Cells by Chitosan-nanoparticles for Translation In Vitro and In Vivo. Mol. Ther.-Nucleic Acids 2014, 3, e173. [Google Scholar] [CrossRef] [PubMed]
- Chahal, J.S.; Khan, O.F.; Cooper, C.L.; Mcpartlan, J.S.; Tsosie, J.K. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. USA 2016, 113, E4133–E4142. [Google Scholar] [CrossRef] [Green Version]
- Chahal, J.S.; Fang, T.; Woodham, A.W.; Khan, O.F.; Ling, J.; Anderson, D.G.; Ploegh, H.L. An RNA nanoparticle vaccine against Zika virus elicits antibody and CD8+T cell responses in a mouse model. Sci. Rep. 2017, 7, 252. [Google Scholar] [CrossRef] [Green Version]
- Démoulins, T.; Ebensen, T.; Schulze, K.; Englezou, P.C.; Pelliccia, M.; Guzmán, C.A.; Ruggli, N.; Mccullough, K.C. Self-replicating RNA vaccine functionality modulated by fine-tuning of polyplex delivery vehicle structure. J. Control. Release 2017, 266, 256–271. [Google Scholar] [CrossRef]
- Englezou, P.C.; Sapet, C.; Démoulins, T.; Milona, P.; Ebensen, T.; Schulze, K.; Guzman, C.; Poulhes, F.; Zelphati, O.; Ruggli, N.; et al. Self-Amplifying Replicon RNA Delivery to Dendritic Cells by Cationic Lipids. Mol. Ther. Nucleic Acid 2018, 12, 118–134. [Google Scholar] [CrossRef]
- Gurnani, P.; Blakney, A.K.; Terracciano, R.; Petch, J.E.; Blok, A.J.; Bouton, R.; Mckay, P.F.; Shattock, R.J.; Alexander, C. The In Vitro, Ex Vivo, and In Vivo Effect of Polymer Hydrophobicity on Charge-Reversible Vectors for Self-Amplifying RNA. Biomacromolecules 2020, 21, 3242–3253. [Google Scholar] [CrossRef]
- Perche, F.; Clemençon, R.; Schulze, K.; Ebensen, T.; Guzmán, C.A.; Pichon, C. Neutral Lipopolyplexes for In Vivo Delivery of Conventional and Replicative RNA Vaccine. Mol. Ther. Nucleic Acid 2019, 17, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Erasmus, J.H.; Khandhar, A.P.; Guderian, J.; Granger, B.; Archer, J.; Archer, M.; Gage, E.; Fuerte-stone, J.; Larson, E.; Lin, S.; et al. A Nanostructured Lipid Carrier for Delivery of a Replicating Viral RNA Provides Single, Low-Dose Protection against Zika. Mol. Ther. 2018, 26, 2507–2522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erasmus, J.H.; Khandhar, A.P.; Connor, M.A.O.; Walls, A.C.; Hemann, E.A.; Murapa, P.; Archer, J.; Leventhal, S.; Fuller, J.T.; Lewis, T.B.; et al. An alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 2020, 12, eabc9396. [Google Scholar] [CrossRef] [PubMed]
- Ajbani, S.P.; Velhal, S.M.; Kadam, R.B.; Patel, V.V.; Bandivdekar, A.H. International Journal of Biological Macromolecules Immunogenicity of Semliki Forest virus based self-amplifying RNA expressing Indian HIV-1C genes in mice. Int. J. Biol. Macromol. 2015, 81, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Blakney, A.K.; Mckay, P.F.; Christensen, D.; Ibarzo, B.; Aldon, Y.; Follmann, F.; Shattock, R.J. Effects of cationic adjuvant formulation particle type, fluidity and immunomodulators on delivery and immunogenicity of saRNA. J. Control. Release 2019, 304, 65–74. [Google Scholar] [CrossRef]
- Biddlecome, A.; Habte, H.H.; Mcgrath, K.M.; Sambanthamoorthy, S.; Wurm, M.; Sykora, M.M.; Knobler, C.M.; Id, I.C.L.; Lasaro, M.; Elbers, K.; et al. Delivery of self-amplifying RNA vaccines in in vitro reconstituted virus-like particles. PLoS ONE 2019, 14, e0215031. [Google Scholar] [CrossRef] [Green Version]
- Kofler, R.M.; Aberle, J.H.; Aberle, S.W.; Allison, S.L.; Heinz, F.X.; Mandl, C.W. Mimicking live flavivirus immunization with a noninfectious RNA vaccine. Proc. Natl. Acad. Sci. USA 2004, 101, 1951–1956. [Google Scholar] [CrossRef] [Green Version]
- Beissert, T.; Koste, L.; Perkovic, M.; Walzer, K.C.; Erbar, S.; Selmi, A.; Diken, M.; Kreiter, S.; Tu, O. Improvement of In Vivo Expression of Genes Delivered by Self-Amplifying RNA Using Vaccinia Virus Immune Evasion Proteins. Hum. Gene Ther. 2017, 28, 1138–1146. [Google Scholar] [CrossRef]
- Lundstrom, K. Self-Amplifying RNA Viruses as RNA Vaccines. Int. J. Mol. Sci. 2020, 21, 5130. [Google Scholar] [CrossRef]
- Moyo, N.; Vogel, A.B.; Buus, S.; Erbar, S.; Wee, E.G.; Sahin, U. Efficient Induction of T Cells against Conserved HIV-1 Regions by Mosaic Vaccines Delivered as Self-Amplifying mRNA. Mol. Ther. Methods Clin. Dev. 2019, 12, 32–46. [Google Scholar] [CrossRef] [Green Version]
- Maruggi, G.; Zhang, C.; Li, J.; Ulmer, J.B.; Yu, D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019, 27, 757–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ly, H.H.; Daniel, S.; Soriano, S.K.V.; Blakney, A.K. Optimization of Lipid Nanoparticles for saRNA Expression and Cellular Activation Using a Design-of-Experiment Approach. Mol. Pharm. 2022, 19, 1892–1905. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef] [PubMed]




| Condition | ClinicalTrials.gov Identifier | Sponsor | Drug Name | Delivery Platform | Administration Route | Status | Completion Date |
|---|---|---|---|---|---|---|---|
| Heart failure | NCT03370887 | AstraZeneca | AZD8601 | Naked mRNA | Epicardial injection | Phase 2 | 30 June 2021 |
| Ulcers associated with T2DM | NCT02935712 | AstraZeneca | AZD8601 | Naked mRNA | Intradermal | Phase 1 | 8 January 2018 |
| Propionic acidemia | NCT04159103 | ModernaTX, Inc. | mRNA-3927 | In vivo/Lipid nano-systems | Intravenous | Phase 1 and 2 | 6 January 2027 |
| Methylmalonic Acidemia | NCT03810690 | ModernaTX, Inc. | mRNA-3704 | In vivo/Lipid nano-systems | Intravenous | Withdrawn | 18 August 2020 |
| OTD | NCT03767270 | Translate Bio, Inc. | MRT5201 | In vivo/Lipid nano-systems | Intravenous | Withdrawn | July 2022 |
| Cystic Fibrosis | NCT03375047 | Translate Bio, Inc. | MRT5005 | In vivo/Lipid nano-systems | Nebulization | Phase 1 and 2 | December 2021 |
| Condition | Location | Sponsors and Collaborators | Estimated Enrollment | Status | Start Date | NCT Number/Phase |
|---|---|---|---|---|---|---|
| COVID-19 | Vietnam | Vinbiocare Biotechnology Joint Stock Company Arcturus Therapeutics, Inc. | 19,400 | Active, not recruiting | August 2021 | NCT05012943 Phase 2/3 |
| Influenza | United States | Pfizer | 468 | Recruiting | February 2022 | NCT05227001 Phase 1 |
| COVID-19 | United States, Singapore | Arcturus Therapeutics, Inc. | 72 | Recruiting | September 2021 | NCT05037097 Phase 1/2 |
| COVID-19 | Uganda | MRC/UVRI and LSHTM Uganda Research Unit | 42 | Recruiting | June 2021 | NCT04934111 Phase 1 |
| COVID-19 | South Africa | ImmunityBio, Inc. | 180 | Recruiting | May 2022 | NCT05370040 Phase 1/2 |
| COVID-19 | United States, Singapore | Arcturus Therapeutics, Inc. | 600 | Terminated | December 2020 | NCT04668339 Phase 2 |
| COVID-19 | United States | HDT Bio Kaiser Permanente Rainier Clinical Research Center C3 Research Associates | 63 | Recruiting | November 2021 | NCT05132907 Phase 1 |
| COVID-19 | United States | GlaxoSmithKline | 10 | Completed | February 2021 | NCT04758962 Phase 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papukashvili, D.; Rcheulishvili, N.; Liu, C.; Ji, Y.; He, Y.; Wang, P.G. Self-Amplifying RNA Approach for Protein Replacement Therapy. Int. J. Mol. Sci. 2022, 23, 12884. https://doi.org/10.3390/ijms232112884
Papukashvili D, Rcheulishvili N, Liu C, Ji Y, He Y, Wang PG. Self-Amplifying RNA Approach for Protein Replacement Therapy. International Journal of Molecular Sciences. 2022; 23(21):12884. https://doi.org/10.3390/ijms232112884
Chicago/Turabian StylePapukashvili, Dimitri, Nino Rcheulishvili, Cong Liu, Yang Ji, Yunjiao He, and Peng George Wang. 2022. "Self-Amplifying RNA Approach for Protein Replacement Therapy" International Journal of Molecular Sciences 23, no. 21: 12884. https://doi.org/10.3390/ijms232112884